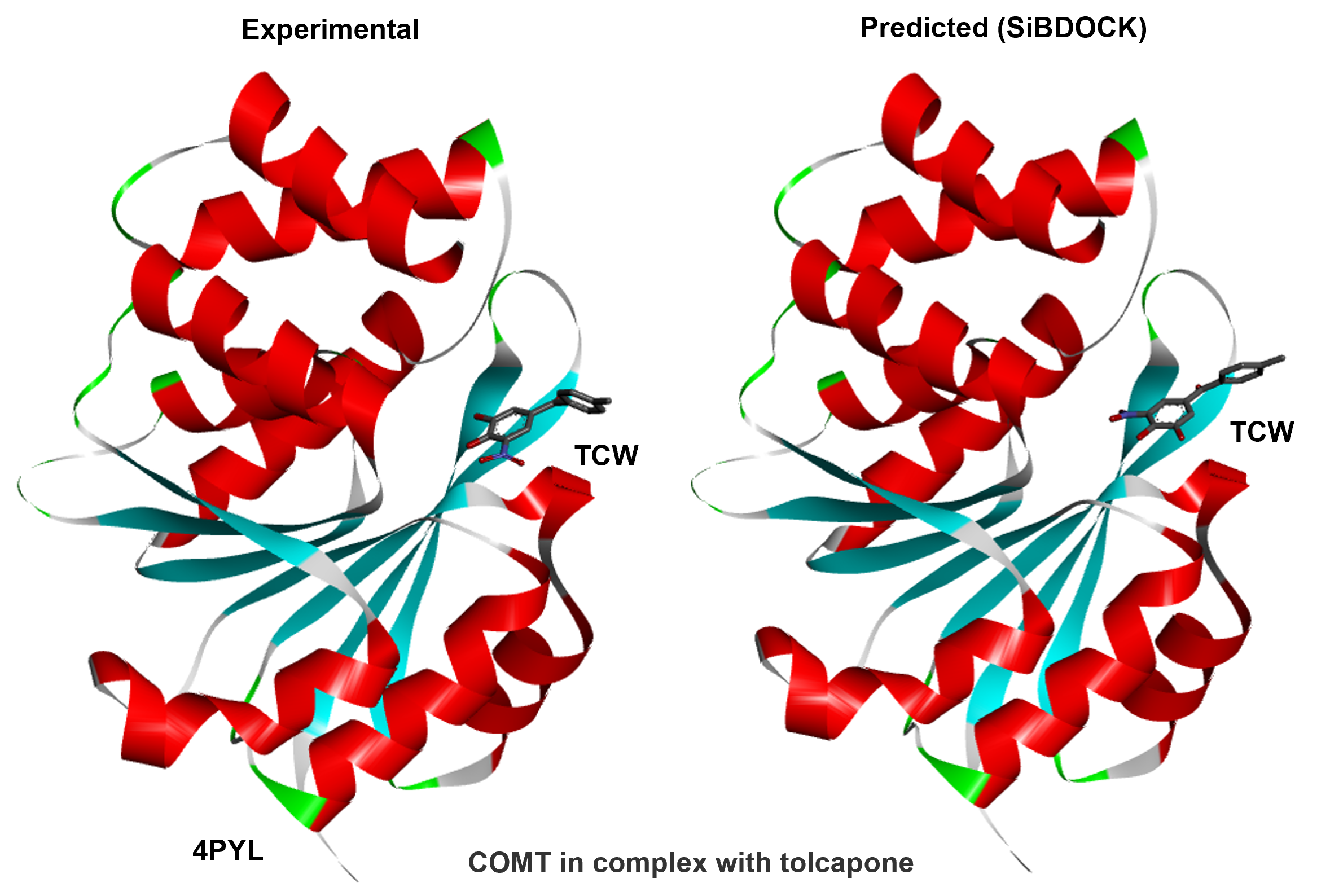
Experience our state-of-the-art novel virtual drug screening technology that aids your rapid drug screening needs.
SiBIOLEAD introduces D-HTVS technology, a user friendly and highly reliable virtual screening technology for virtual screening small molecule libraries with millions of compounds.
Docking large chemical libraries containing millions of compounds against target protein is a hassle process and requires large computational power.
The D-HTVS technology predicts avid binding small molecules with variety of scaffolds along with their structurally-related molecules against your target protein.
D-HTVS method follows two stage virtual screening procedure for your target macromolecules:
1) First stage virtual docking covers the entire chemical space (diversity/scaffolds) in the target library, e.g., ChEMBL, and ranked based on the docking scores.
2) All the structurally related compounds for the top 10 predicted diverse set will be fetched from the target library and will be docked against your target macromolecules.
Finally results will be generated and ranked based on docking scores.
Our D-HTVS technology reduces the computational time for screening humongous libraries significantly. Typical runtime for 800,000 compounds in D-HTVS mode is estimated to be 12-15 hours, which by traditional screening may take more than 5 to 6 days in a 32-core processor machine.
For users convenience, SiBIOLEAD offers widely used chemical libraries as part of D-HTVS module to ease the virtual drug-discovery process. Therefore no need for pre-processing millions of ligands.
ChemBridge is one of the leading global providers of enabling chemistry products and contract research services for small molecule drug discovery for more than two decades. ChemBridge portfolio contains more than 1.2 million small molecules. For hit finding across different targets, against novel targets or for discovery of new chemical series against existing targets.
ChemBridge molecules have been used across many disease areas resulting in hundreds of citations in published research articles. Click here for more details
ChEMBL is a curated database of bioactive drug-like small molecules based on calculated properties, including logP, Molecular Weight, Lipinski rule. ChEMBL database contains over 2 million compounds for screening.
ZINC is a free commercially-available database of compounds for virtual screening. ZINC contains over 230 million purchasable compounds in ready-to-dock, 3D formats. For our users convenience, we are providing ZINC natural-product subsets , as a pre-loaded library.
Open NCI database is an openly accessible database with the computer structures being made available by DTP as public domain data. Currenlty there are ~ 265,000 compounds available in open NCI database.
Our High-throughput Virtual Screening (HTVS) module is an interactive HTVS job submission module for AutoDock-VINA. Users can dock upto 5000 molecules per submission. Our in-house developed HPC technology gears up the docking calculation in high throughput (low exhaustive) mode with a typical runtime rate of 4000 molecules/hour excluding the pre-processing step.
SiBioLead offers an interactive results page including 3D molecular viewer that allow users to view the protein-ligand complex in 3D environment. We use JSMOL open source 3D viewer for rendering protein-ligand complex. Submitted ligands will be ranked based on their interaction energies with the target protein.
In addition to protein-ligand docking, SiBioLead platform also performs protein-ligand interaction analysis using Protein ligand Interaction Profiler (PLIP) software package.